4. Genome assembly¶
4.1. Preface¶
In this section we will use our skill on the command-line interface to create a genome assembly from sequencing data.
Note
You will encounter some To-do sections at times. Write the solutions and answers into a text-file.
4.2. Overview¶
The part of the workflow we will work on in this section can be viewed in Fig. 4.1.
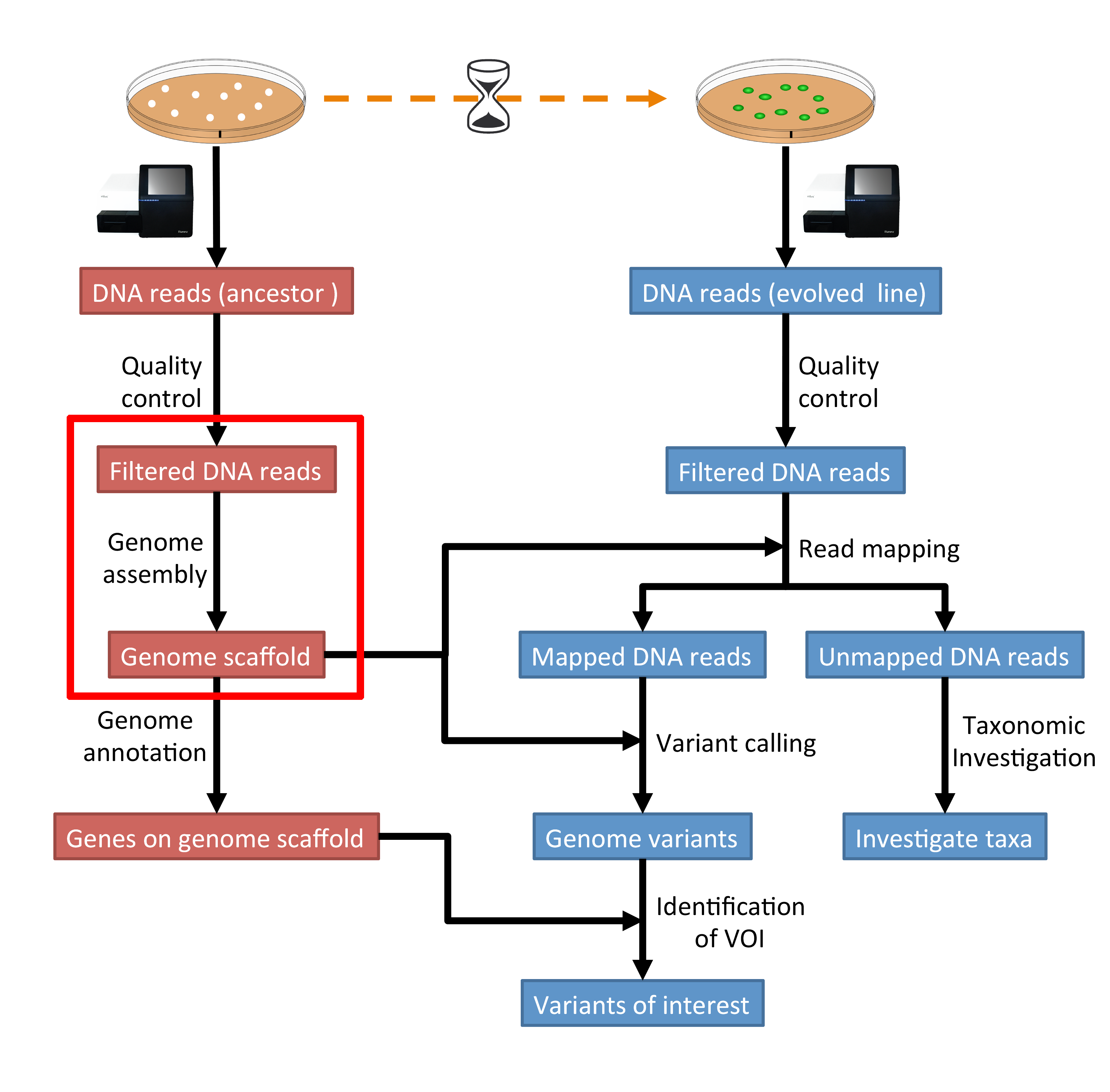
Fig. 4.1 The part of the workflow we will work on in this section marked in red.¶
4.3. Learning outcomes¶
After studying this tutorial you should be able to:
Compute and interpret a whole genome assembly.
Judge the quality of a genome assembly.
4.4. Before we start¶
Lets see how our directory structure looks so far:
$ cd ~/analysis
$ ls -1F
data/
multiqc_data/
multiqc_report.html
trimmed/
trimmed-fastqc/
Attention
If you have not run the previous section Quality control, you can download the trimmed data needed for this section here: Downloads. Download the file to the ~/analysis directory and decompress. Alternatively on the CLI try:
cd ~/analysis
wget -O trimmed.tar.gz https://osf.io/m3wpr/download
tar xvzf trimmed.tar.gz
4.5. Creating a genome assembly¶
We want to create a genome assembly for our ancestor. We are going to use the quality trimmed forward and backward DNA sequences and use a program called SPAdes to build a genome assembly.
Todo
Discuss briefly why we are using the ancestral sequences to create a reference genome as opposed to the evolved line.
4.5.1. Installing the software¶
We are going to use a program called SPAdes fo assembling our genome. In a recent evaluation of assembly software, SPAdes was found to be a good choice for fungal genomes [ABBAS2014]. It is also simple to install and use.
$ conda create -n assembly spades quast
$ conda activate assembly
4.5.2. SPAdes usage¶
# change to your analysis root folder
$ cd ~/analysis
# first create a output directory for the assemblies
$ mkdir assembly
# to get a help for spades and an overview of the parameter type:
$ spades.py -h
Generally, paired-end data is submitted in the following way to SPAdes:
$ spades.py -o result-directory -1 read1.fastq.gz -2 read2.fastq.gz
Todo
Run SPAdes with default parameters on the ancestor’s trimmed reads
Read in the SPAdes manual about about assembling with 2x150bp reads
Run SPAdes a second time but use the options suggested at the SPAdes manual section 3.4 for assembling 2x150bp paired-end reads. Use a different output directory
assembly/spades-150for this run.
Hint
Should you not get it right, try the commands in Code: SPAdes assembly (trimmed data).
4.6. Assembly quality assessment¶
4.6.1. Assembly statistics¶
Quast (QUality ASsessment Tool) [GUREVICH2013], evaluates genome assemblies by computing various metrics, including:
N50: length for which the collection of all contigs of that length or longer covers at least 50% of assembly length
NG50: where length of the reference genome is being covered
NA50 and NGA50: where aligned blocks instead of contigs are taken
miss-assemblies: miss-assembled and unaligned contigs or contigs bases
genes and operons covered
It is easy with Quast to compare these measures among several assemblies. The program can be used on their website.
$ conda install quast
Run Quast with both assembly scaffolds.fasta files to compare the results.
$ quast -o assembly/quast assembly/spades-default/scaffolds.fasta assembly/spades-150/scaffolds.fasta
Todo
Compare the results of Quast with regards to the two different assemblies.
Which one do you prefer and why?
4.7. Compare the untrimmed data¶
Todo
To see if our trimming procedure has an influence on our assembly, run the same command you used on the trimmed data on the original untrimmed data.
Run Quast on the assembly and compare the statistics to the one derived for the trimmed data set. Write down your observations.
Hint
Should you not get it right, try the commands in Code: SPAdes assembly (original data).
4.8. Further reading¶
4.8.1. Background on Genome Assemblies¶
How to apply de Bruijn graphs to genome assembly. [COMPEAU2011]
Sequence assembly demystified. [NAGARAJAN2013]
4.8.2. Evaluation of Genome Assembly Software¶
GAGE: A critical evaluation of genome assemblies and assembly algorithms. [SALZBERG2012]
Assessment of de novo assemblers for draft genomes: a case study with fungal genomes. [ABBAS2014]
4.9. Web links¶
Lectures for this topic: Genome Assembly: An Introduction
Bandage (Bioinformatics Application for Navigating De novo Assembly Graphs Easily) is a program that visualizes a genome assembly as a graph [WICK2015].
References
- ABBAS2014(1,2)
Abbas MM, Malluhi QM, Balakrishnan P. Assessment of de novo assemblers for draft genomes: a case study with fungal genomes. BMC Genomics. 2014;15 Suppl 9:S10. doi: 10.1186/1471-2164-15-S9-S10. Epub 2014 Dec 8.
- COMPEAU2011
Compeau PE, Pevzner PA, Tesler G. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol. 2011 Nov 8;29(11):987-91
- GUREVICH2013
Gurevich A, Saveliev V, Vyahhi N and Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013, 29(8), 1072-1075
- NAGARAJAN2013
Nagarajan N, Pop M. Sequence assembly demystified. Nat Rev Genet. 2013 Mar;14(3):157-67
- SALZBERG2012
Salzberg SL, Phillippy AM, Zimin A, Puiu D, Magoc T, Koren S, Treangen TJ, Schatz MC, Delcher AL, Roberts M, Marçais G, Pop M, Yorke JA. GAGE: A critical evaluation of genome assemblies and assembly algorithms. Genome Res. 2012 Mar;22(3):557-67
- WICK2015
Wick RR, Schultz MB, Zobel J and Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 2015, 10.1093/bioinformatics/btv383