1. Introduction¶
This is an introductory tutorial for learning genomics mostly on the Linux command-line. Should you need to refresh your knowledge about either Linux or the command-line, have a look here.
In this tutorial you will learn how to analyse next-generation sequencing (NGS) data. The data you will be using is actual research data. The experiment follows a similar strategy as in what is called an “experimental evolution” experiment [KAWECKI2012], [ZEYL2006]. The final aim is to identify the genome variations in evolved lines of E. coli that can explain the observed biological phenotype(s).
1.1. The workflow¶
The tutorial workflow is summarised in Fig. 1.1.
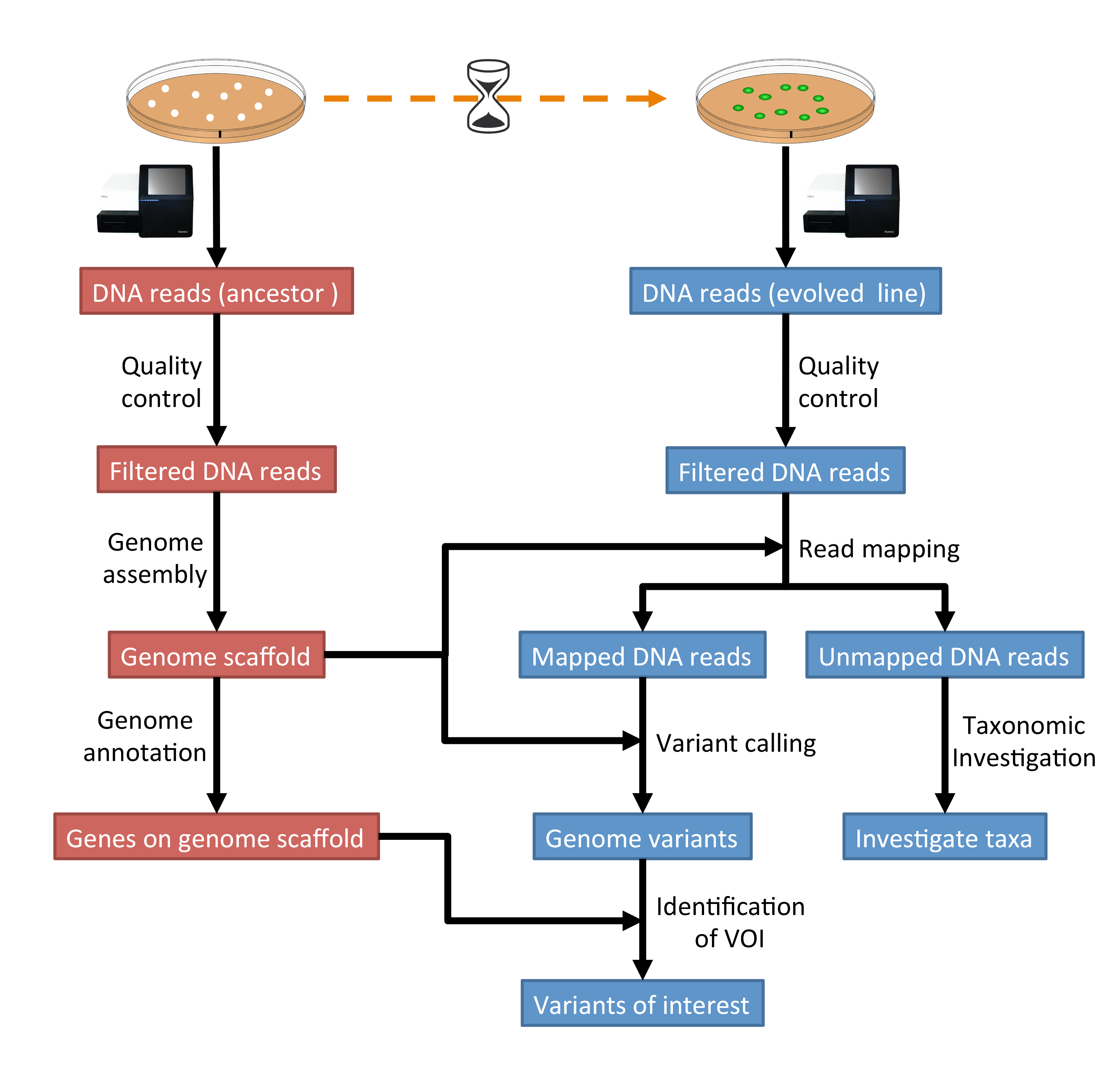
Fig. 1.1 The tutorial will follow this workflow.¶
1.2. Learning outcomes¶
During this tutorial you will learn to:
Check the data quality of an NGS experiment
Create a genome assembly of the ancestor based on NGS data
Map NGS reads of evolved lines to the created ancestral reference genome
Call genome variations/mutations in the evolved lines
Annotate a newly derived reference genome
Find variants of interest that may be responsible for the observed evolved phenotype(s)
References
- KAWECKI2012
Kawecki TJ et al. Experimental evolution. Trends in Ecology and Evolution (2012) 27:10
- ZEYL2006
Zeyl C. Experimental evolution with yeast. FEMS Yeast Res, 2006, 685–691